The Disease
T-cell acute lymphoblastic leukemia: an unresolved epigenetic disease
Up to 25% of children with Acute Lymphoblastic Leukemia will fail frontline therapy and their prognosis is dismal. Cure rates for patients relapsing on therapy are approximate 20%. While patients with later relapses have a better prognosis, the majority eventually succumbs to the disease (overall cure rate 40 to 50%). There have been several obstacles to the successful treatment of relapsed ALL like lower remission-reinduction rates and early second relapse for those who enter remission. The actual clinical issue of acute leukemia is survival after relapse. Many biological and clinical variables correlate with outcome in newly diagnosed patients, but few variables predict outcome for relapsed disease. Only site and timing of relapse, as well as blast immunophenotype, have consistently emerged as prognostic variables in relapsed ALL. The earlier relapse occurs relative to the time of initial diagnosis, the worse the outcome. Whereas 40 to 50% of children with “late” marrow relapse, defined as occurring at least 3 years from initial diagnosis are cured, less than 10% of children survive relapse that occurs within the first year from diagnosis. The site of recurrence has also been shown to be of prognostic significance. Finally, as the intensity of frontline therapy for ALL has increased, the question of whether recurrent disease has become more difficult to treat has been raised.
Experimental System and Technologies
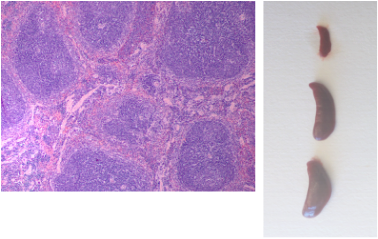 Analysis of mice for leukemia progression: Hematoxylin/Eosin staining of the spleen for detection of leukemic infiltration (UP). Comparison of spleen size amongst a healthy mouse (top) and two leukemic mice (middle and bottom) (BOTTOM).
Analysis of mice for leukemia progression: Hematoxylin/Eosin staining of the spleen for detection of leukemic infiltration (UP). Comparison of spleen size amongst a healthy mouse (top) and two leukemic mice (middle and bottom) (BOTTOM).
The Ntziachristos laboratory studies the mechanistic aspects of oncogenesis with an emphasis on transcriptional and epigenetic regulation of acute leukemia. Important questions are related to how oncogenes interact with each other and with epigenetic modulators to influence gene expression programs as well as how their function is related to three-dimensional (3D) structure of the nucleus, how oncogenes are control at the post-translational level via (de)ubiquitination and other biological aspects of cancer cells, such as splicing.
To address these questions we use high-throughput molecular and cell biology techniques like ChIP-Seq, RNA-Seq, 4C-Seq and HiC, fluorescent in situ hybridization, biochemical analysis e.t.c. in cell lines and cells of human origin and tissues of mouse models of disease. We also employ biochemical technologies such as ubiquitination analysis (TUBE, ubiquitin pull-downs), gel filtration, immune-precipitations and proteomics analysis. We have also employed xenograft models including the use of luciferase-expressing cell lines and patient cells. In addition to understanding cancer biology these findings help us design and test targeted therapies in preclinical models of leukemia.
To address these questions we use high-throughput molecular and cell biology techniques like ChIP-Seq, RNA-Seq, 4C-Seq and HiC, fluorescent in situ hybridization, biochemical analysis e.t.c. in cell lines and cells of human origin and tissues of mouse models of disease. We also employ biochemical technologies such as ubiquitination analysis (TUBE, ubiquitin pull-downs), gel filtration, immune-precipitations and proteomics analysis. We have also employed xenograft models including the use of luciferase-expressing cell lines and patient cells. In addition to understanding cancer biology these findings help us design and test targeted therapies in preclinical models of leukemia.
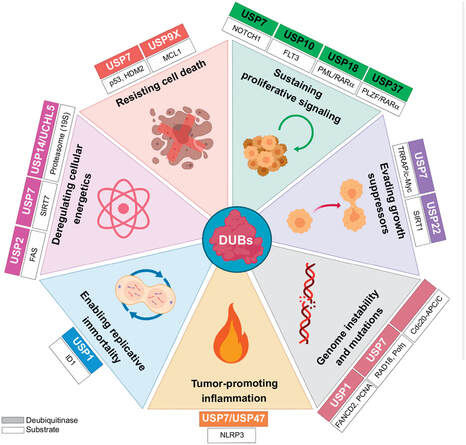
Aspects of Leukemia Biology Influenced by Deubiquitinases (DUBs)
The different DUBs can maintain an oncogenic phenotype in leukemia through the stabilization, activation, and relocalization of their targets. Abbreviations: USP, ubiquitin-specific protease; RARα, retinoic acid receptor alpha; PLZF, promyelocytic leukemia zinc finger; TRRAP, transformation/transcription domain associated protein; SIRT, sirtuin; ID1, inhibitor of DNA binding 1.
The different DUBs can maintain an oncogenic phenotype in leukemia through the stabilization, activation, and relocalization of their targets. Abbreviations: USP, ubiquitin-specific protease; RARα, retinoic acid receptor alpha; PLZF, promyelocytic leukemia zinc finger; TRRAP, transformation/transcription domain associated protein; SIRT, sirtuin; ID1, inhibitor of DNA binding 1.
Ghent UNIVERSITY |
The laboratory is a member of the Department of Biololecular Medicine situated in the University Hospital area in small distance from Ghent city center. The group has access to cutting-edge technology and facilities and is exposed to the vibrant scientific community of Ghent University and other research centers in the Ghent area and Belgium. Scientists in the group have the opportunity to present their work and get feedback in Institutional, National and International forums.
|
